Modelling parasite-produced marine diseases - The case of the mass mortality event of Pinna nobilis
Introduction
The noble fan mussel (Pinna nobilis) is the largest endemic bivalve in the Mediterranean Sea, where it plays a crucial ecological role in its habitat by contributing to water clarity, being a habitat-forming species and even working as ecosystem engineers, creating biogenic reefs. Nowadays, P. nobilis is under a serious extinction risk due to a Mass Mortality Event (MME) that has occurred throughout the whole Mediterranean basin very recently. The main cause of this mortality is the protozoan Haplosporidium pinnae, which is spread by marine currents through the Mediterranean basin causing an epidemic (M. Cabanellas-Reboredo 2019; G. Catanese 2018).
Contact and vector-borne based infectious diseases of terrestrial vertebrates and their epidemiology are typically studied using variations of the classical formulation of the Kermack and McKendrick SIR model. However, the state of the art of epidemiological studies in marine ecosystems lags behind that of their terrestrial counterparts. In fact, compartmental models are starting to be used only recently in the study of marine epizootics, in particular in bivalve epidemics (G. Bidegain 2016a, G. Bidegain 2016b, G. Bidegain 2017).
In the present work we analyse a model that is aimed to describe disease transmission from an infected immobile host to a susceptible one of the same species through waterborne parasites, that are explicitly described. The model is closely related to the SIP model introduced in (G. Bidegain 2016b). In this first study we analyse in detail the properties of the mean-field version of the model, that aims to describe spatially homogeneous (i.e. well mixed) populations.
In the following sections I’ll try to explain the model and the main results of the study in a nontechnical and pedagogical way. For those further interested, just read the paper!
The model
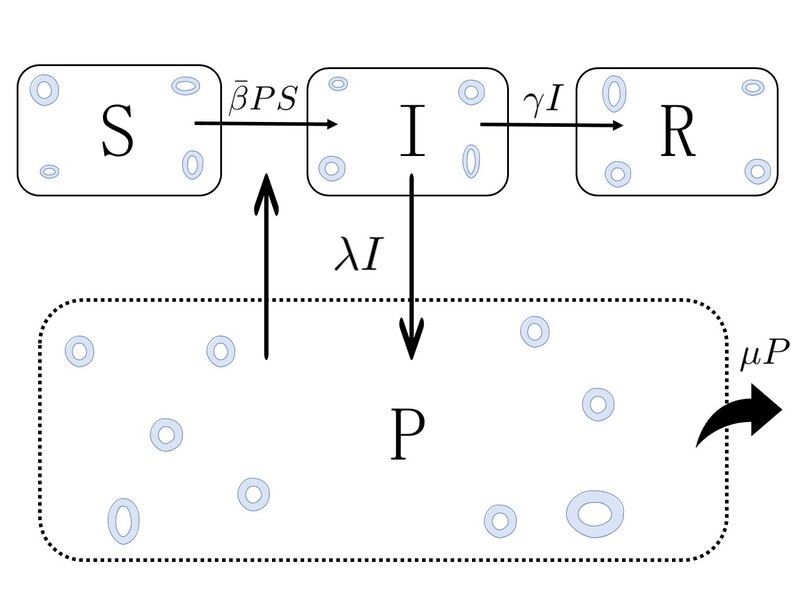
The model is defined according to the diagram above. We consider the parasite population in the marine medium (\(P\)) and the hosts population, that can be in three diferent states: susceptible (\(S\)), infected by the parasite (\(I\)) and dead (\(R\)). Then, we consider the different epidemic processes that can occur in this pathosystem: susceptible hosts can get infected after filtering parasites, infected hosts can die, infected hosts can produce parasites and release them in the medium and parasites can die. Each of these processes will occur at different times, ones more than the other, etc. This will be controled through the parameters of the model, which are nothing but the probability per unit time (rate) that each of this processes take place. As can be observed in the diagram, \(\beta\) is the rate at which susceptible hosts are infected upon filtering parasites, \(\gamma\) is the rate at which infected hosts die, \(\lambda\) is the rate at which infected hosts produce parasites and \(\mu\) is the rate at which parasites die. Altogether, and after some assumptions and considerations, one can write a mathematical model that describes the time-evolution of the number of hosts in each state and the number of parasites in the system,
\[\begin{aligned} \dot{S} &=-\bar{\beta} P S \\ \dot{I} &=\bar{\beta} P S-\gamma I \\ \dot{R} &=\gamma I \\ \dot{P} &=\lambda I-\bar{\beta} P S-\mu P \ . \end{aligned}\]More formally, this kind of models are known as compartmental models and the mathematical framework we are using to represent it is known as a system of ordinary differential equations.
Results
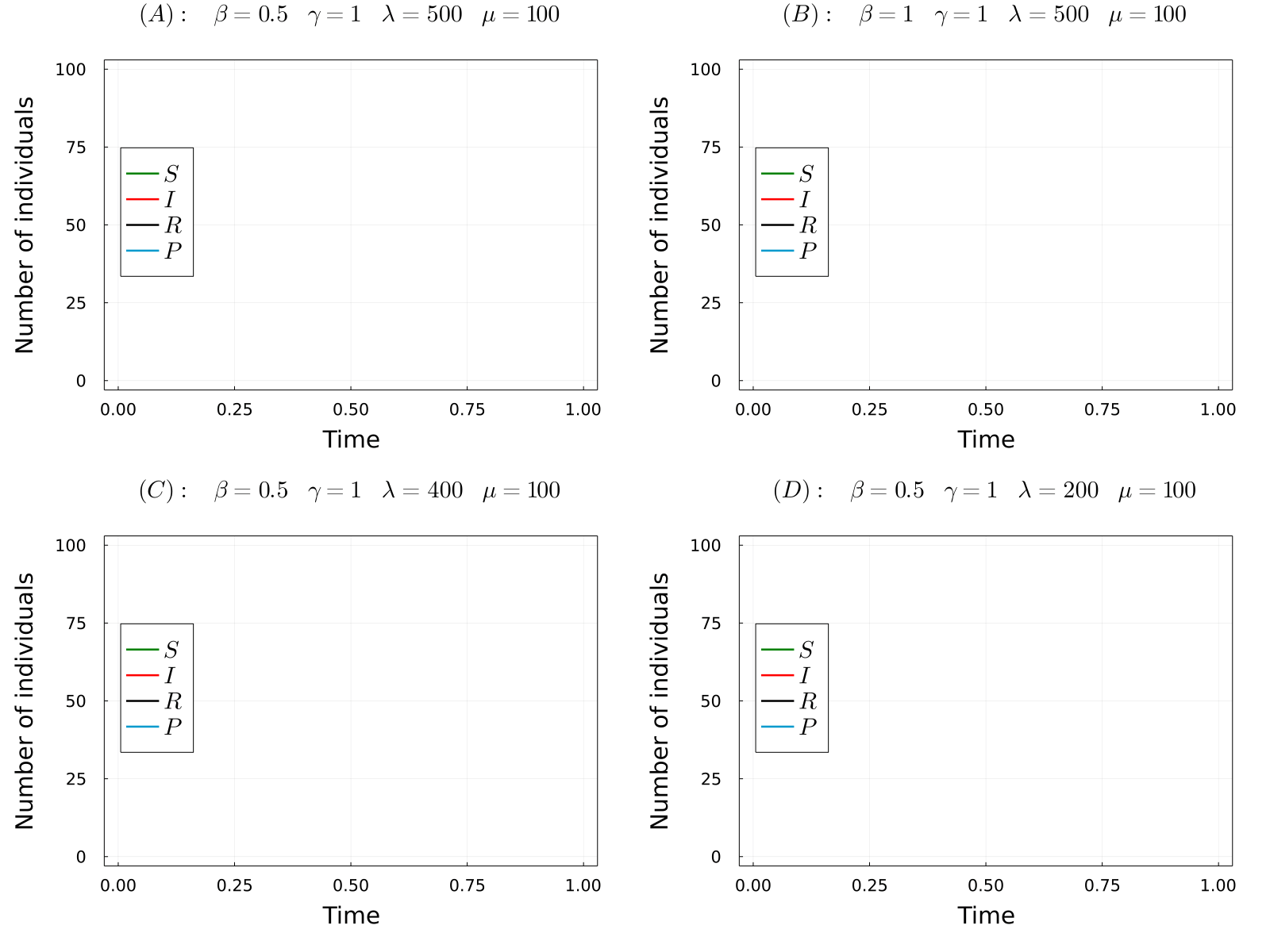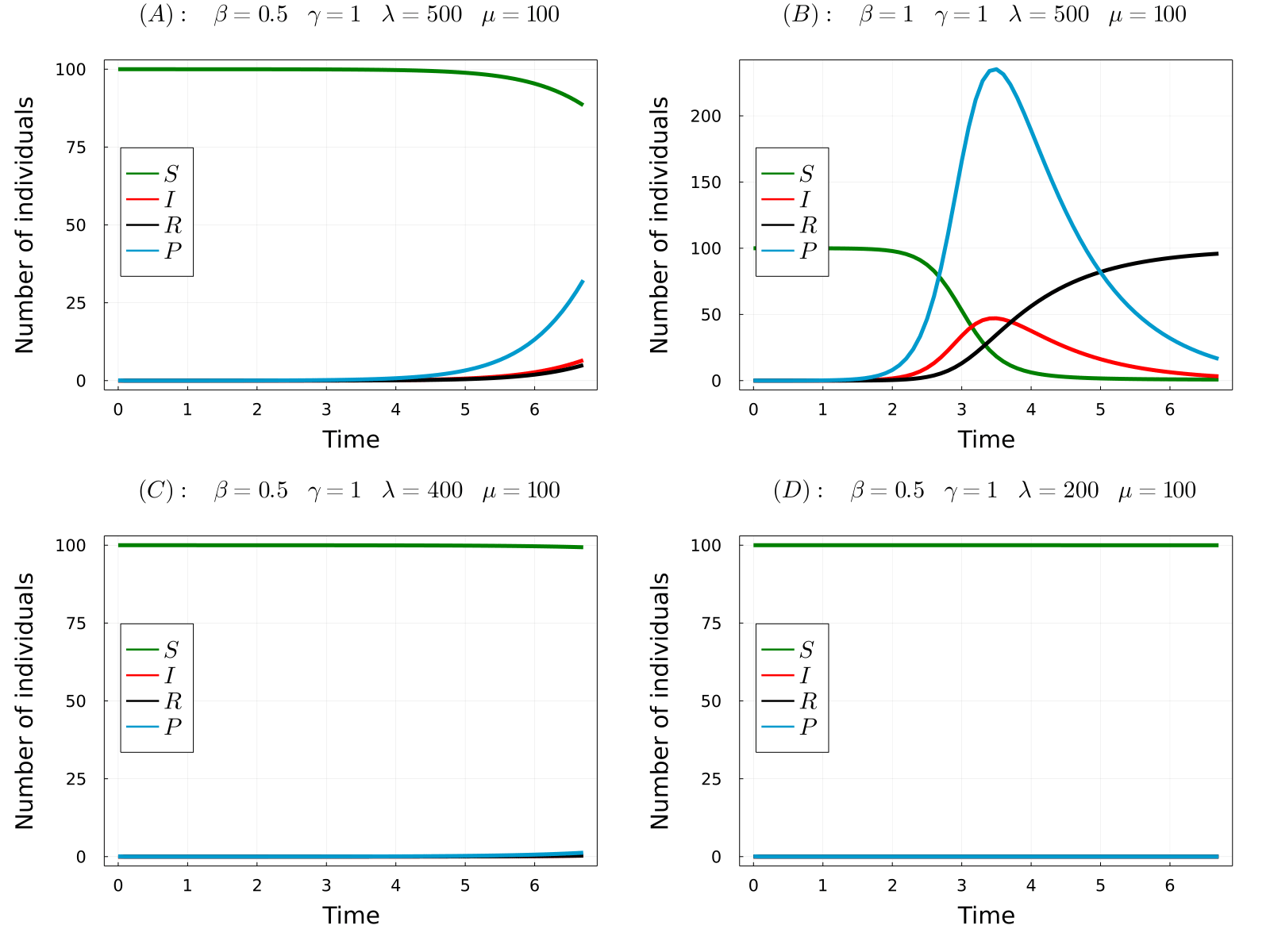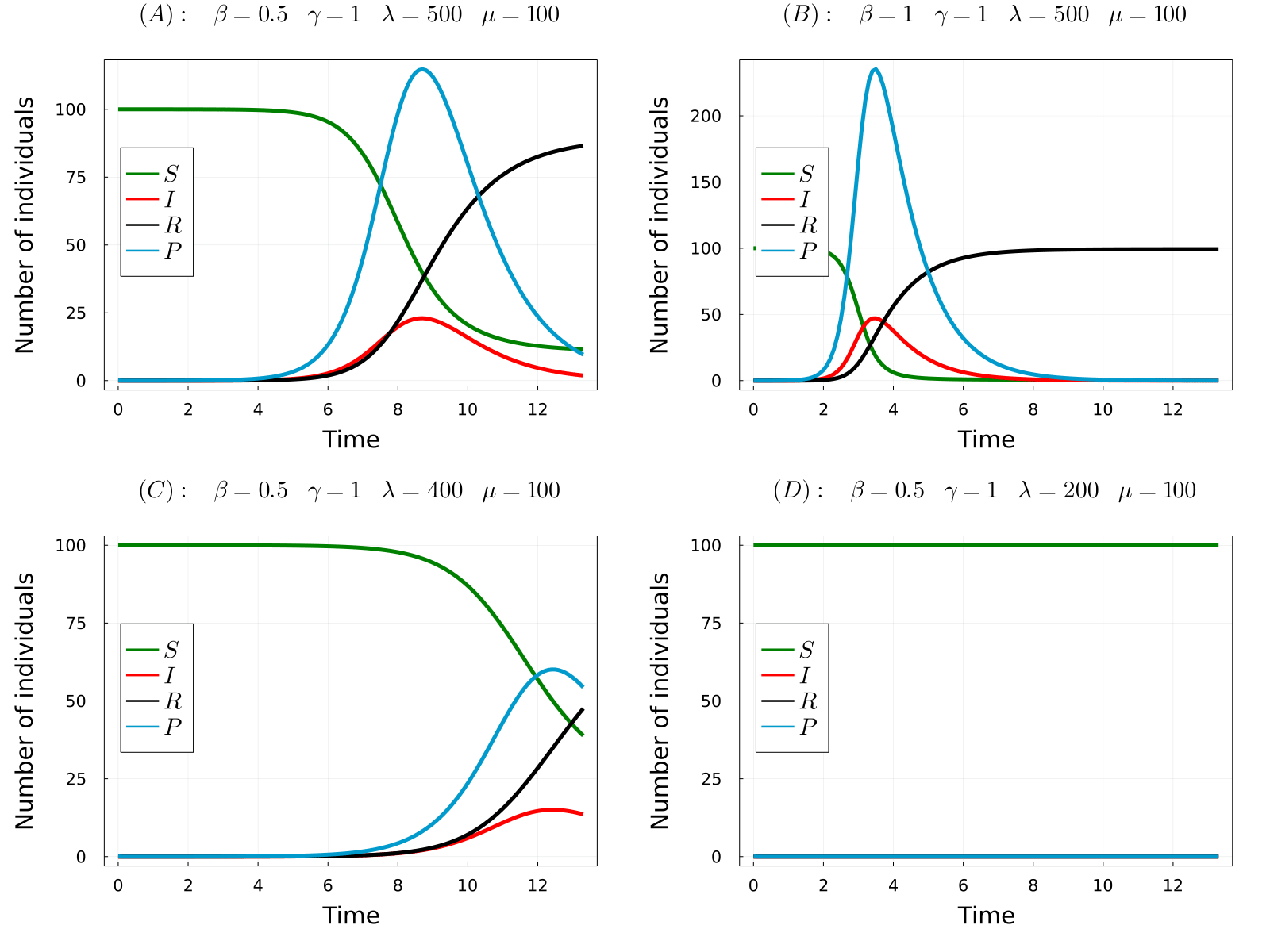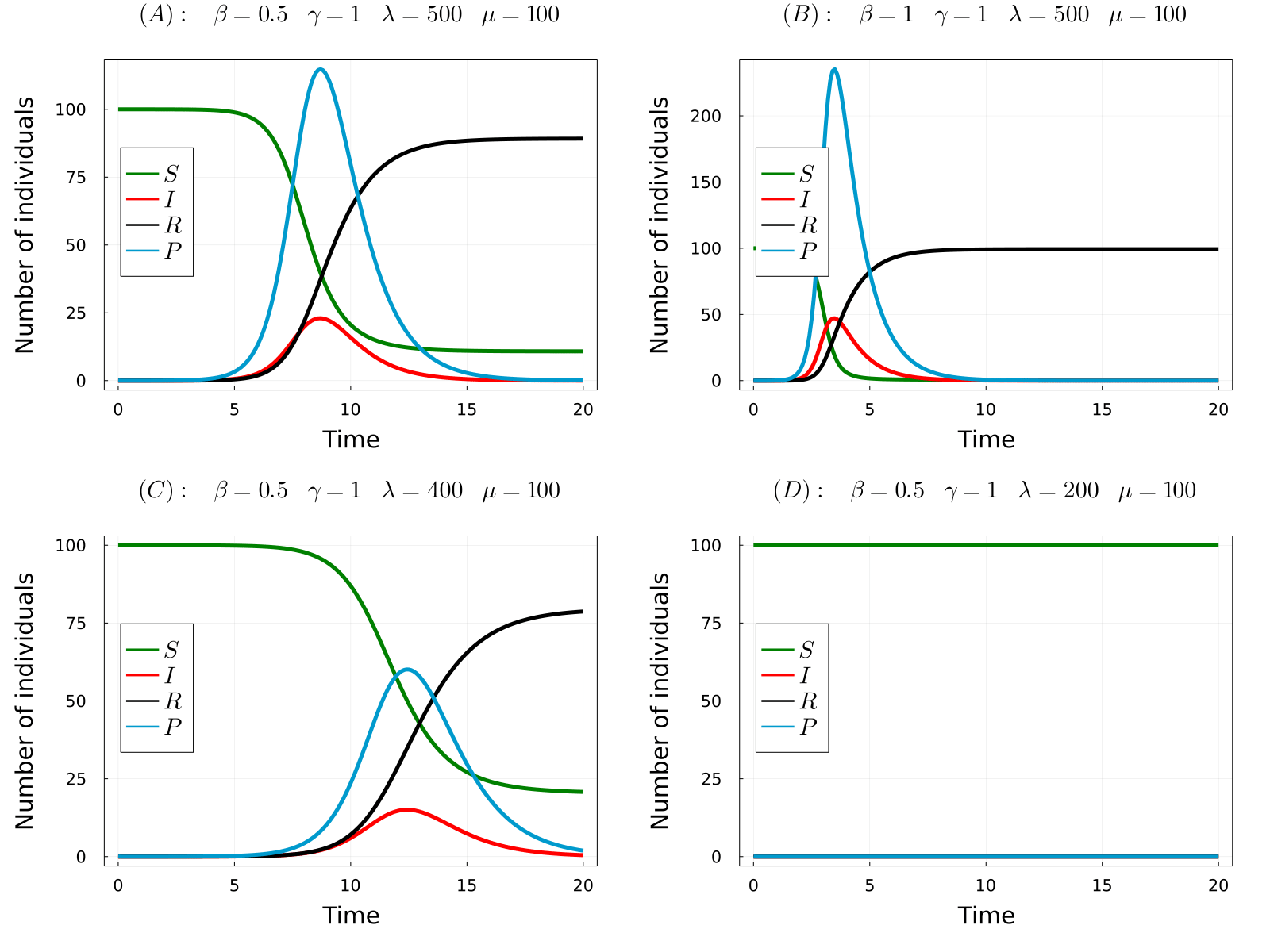
The developed model can be used to simulate the considered processes for different values of the parameters and obtain the time-evolution of the number of hosts in each state and the number of parasites in the system. In the gif below we show the results of the model for four different parameters sets, from which we can make a number of observations. Comparing (A) with (B), we observe that increasing the transmission rate (\(\beta\)) the epidemic starts earlier and the final number of dead hosts increase, something that indeed could be expected from common sense. Similarly, comparing plots (A) and (C) we note that decreasing the parasite production rate of infected hosts (\(\lambda\)) has the opposite effects, causes a delay in the appearence of the epidemic and reduces the final number of dead hosts at the end of the epidemic. Again, this behaviour could be anticipated from common sense. Finaly, we observe that if we further reduce the \(\lambda\) parameter (the same would happen with \(\beta\)), no outbreak is produced.
One could further study the pathosystem by changing the parameter values of the model and observing the simulation outcome, but this would be tedious and inefficient. Instead, we can use some mathematical theories from dynamical systems to obtain something much more interesting: the so-called Basic Reproduction Number, \(R_0\). In short, this parameter measures the average number of secondary infections produced by an initial infected individual in a fully susceptible population. So, if \(R_0>1\), an infected host produces more than one new infection, giving rise to a branching process that causes the exponential rise of infected cases. On the other hand, if \(R_0<1\) an infected host infects causes less than one new infection and the epidemic dies out. For our model, the Basic Reproduction Number is given by the following formula:
\[R_0=\frac{\lambda}{\gamma\left(1+\displaystyle\frac{\mu}{\beta}\right)}\]As you can see, the result is pretty simple! Now we can check the \(R_0\) values of the previous simulations, which turn out to be:
- (A) \(R_0\approx2.5\)
- (B) \(R_0\approx5\)
- (C) \(R_0\approx2\)
- (D) \(R_0\approx0.995\)
Note that in the only case that \(R_0<1\), this is case (D), an outbreak was not developed in the simulations. Thus, the theory perfectly predicts the appearence of an outbreak. So now we can fully understand the behaviour of the model without even simulating it! Indeed, it can be proven that the number of dead individuals at the end of the epidemic is a (more complicated) function of \(R_0\), so that only by knowing the parameter values of the model you can even predict the consequences of the epidemic
Up to this point I hope to have convinced you of the power of applying mathematics to study epidemics, at least this one. But, all this still seems very theoretical, so, it is really useful? Well, it all depends on the knowledge of the real values of the parameters and the availability of field data. For the case of the Mass Mortality Event of Pinna nobilis, as it is an emergent disease, only the value of the mortality rate of hosts could be inferred, being \(\gamma=1\) month\(^{-1}\). However, some data of the time evolution of the epidemic in the controlled water tanks was available after a frustrated conservation effort. The empirical data consists of the proportion of survivors as a function of time in the controlled water tanks with a temporal resolution of one month. The availability of such data allowed us to fit our model, this is, obtain the values of the model parameters that better represents the data.
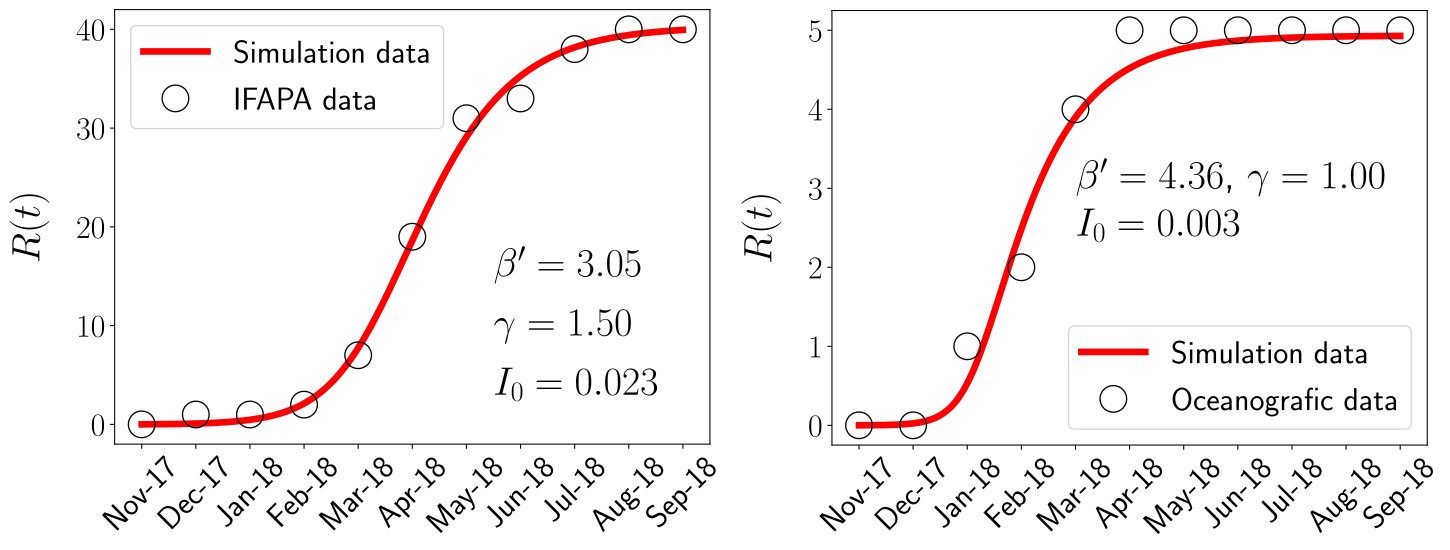
Indeed, the fitting procedure involved further mathematical analysis that is out of the scope of this outreach article. In short, we reduced our model to a simpler (less equations and less parameters) but equivalent one, that turn out to be a well-known epidemic model, the SIR model. In any case, in the figure above we can observe how the model (in its reduced version) is able to chracterize the outbreaks on two different water tanks, that were kept at different temperatures.
Conclusions
In this study, we developed a deterministic compartmental model to describe parasite produced marine diseases of immobile hosts and applied it to the particular case of the Mass Mortality Event of Pinna nobilis. The results presented here represent only the surface of the study, if you are further interested go to read the paper!
References
Cabanellas-Reboredo M., et al. Tracking a mass mortality outbreak of pen shell Pinna nobilis populations: A collaborative effort of scientists and citizens Sci. Rep., 9 (2019)
Catanese G., et al. Haplosporidium pinnae sp. nov. a haplosporidan parasite associated with mass mortalities of the fan mussel, Pinna nobilis, in the Western Mediterranean Sea J. Invertebr. Pathol., 157 (2018)
Bidegain G., Powell E., Klinck J., Ben-Horin T., Hofmann E. Microparasitic disease dynamics in benthic suspension feeders: Infective dose, non-focal hosts, and particle diffusion Ecol. Model., 328 (2016)
Bidegain G., Powell E.N., Klinck J.M., Ben-Horin T., Hofmann E.E. Marine infectious disease dynamics and outbreak thresholds: contact transmission, pandemic infection, and the potential role of filter feeders Ecosphere, 7 (2016).
Bidegain G., Powell E., Klinck J., Hofmann E., Ben-Horin T., Bushek D., Ford S., Munroe D., Guo X. Modeling the transmission of Perkinsus marinus in the eastern oyster Crassostrea virginica Fish. Res., 186 (2017)




